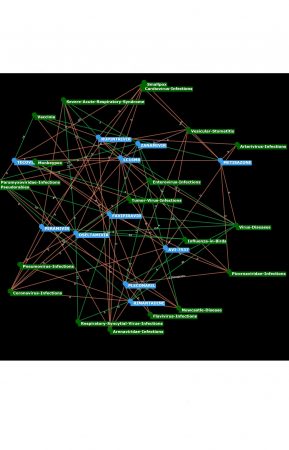
Drug-Disease-Networks – New innovative PubPharm-Tool
Recent artificial intelligence (AI) models can be used to automatically detect existing associations between drugs and diseases, as well as predict new associations based on literature. However, in many cases the explainability of an AI-based prediction is difficult to comprehend, and thus poses an interpretation problem. A possible solution to this problem is to explain such predictions implicitly using other drugs and diseases, e.g., a chemically similar drug is also likely to have similar properties, and thus likely to have similar and known drug-disease associations. The known drug-disease associations can help explain a predicted association. Network views (e.g. EMBL STITCH) are a powerful tool to explore such complex pharmaceutical associations. A network view in which the individual pharmaceutical entities are connected to each other via associations enables a quick exploration of the known and predicted drug-disease associations.
In this context, a new innovative PubPharm function – Drug-Disease-Networks – was developed by our colleague Janus Wawrzinek from the Institute for Information Systems. These Drug-Disease-Networks are now available in a BETA version in PubPharm. With the help of AI (in detail using neural networks), relationships between pharmacy-relevant entities (substances, diseases, and genes) were determined based on research literature. The drug-disease networks complement and visualise the suggestion function in PubPharm, which provides contextually similar, related substances, diseases/symptoms, and genes when searching for diseases or active substances. The following could be determined in an online survey (N=157) regarding the use of the suggestion function: Approximately 70% of the people who use PubPharm several times a month or more stated that they use this context-based information (semantic faceting) for substances, diseases/symptoms or genes several times a month or more.
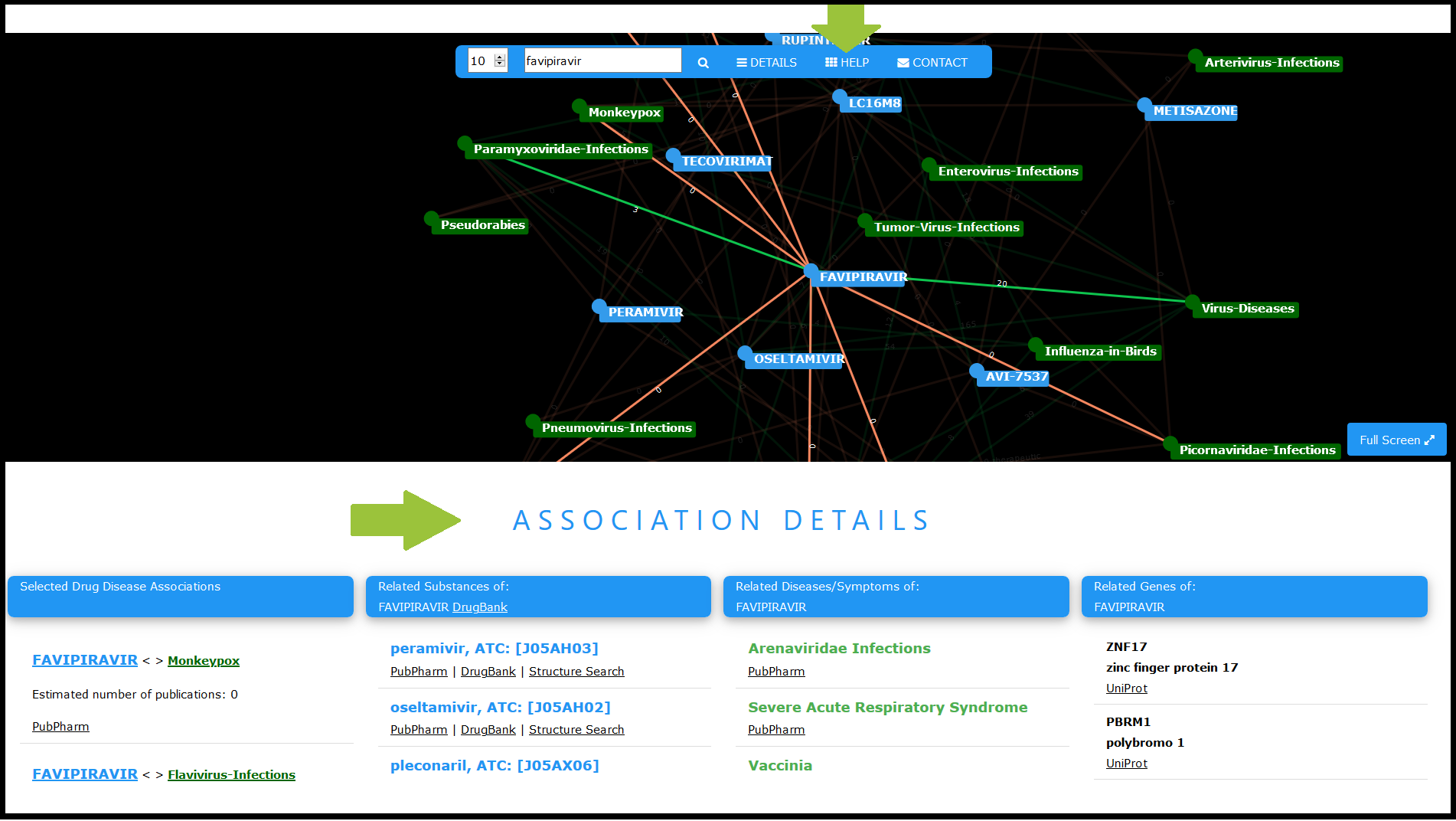
In PubPharm, you can get access to the Drug-Disease-Networks below the central search box. Detailed explanations on usability are provided in the integrated help function.
Links to further information (e.g. to the Comparative Toxicogenomics Database and to the DrugBank database) support the interpretation of the sometimes complex entity associations.
Your feedback will help us to further optimise and adapt this innovative tool to your needs. Please send us comments and suggestions via e-mail to pubpharm[at]tu-braunschweig.de. Thank you for your support!